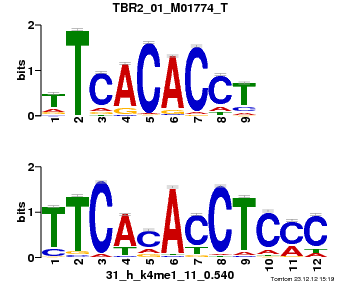
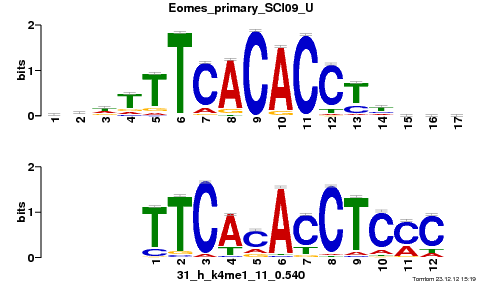
Summary
|
Alignment
|
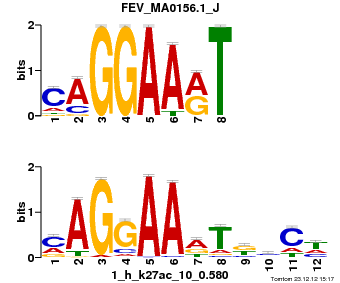
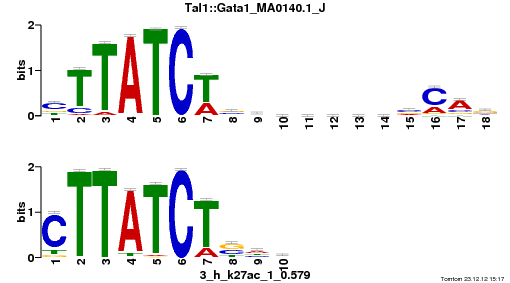
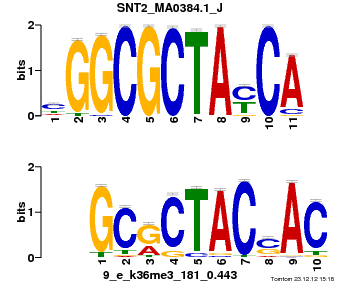
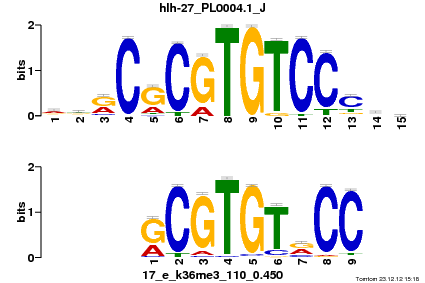
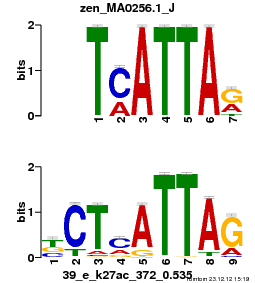
| Name |
Tal1::Gata1_MA0140.1_J |
| Database |
trnsfcJsprUnprobHpdiCmbned3.meme |
|
p-value |
1.4808e-08 |
|
E-value |
4.3254e-05 |
|
q-value |
8.63029e-05 |
| Overlap |
10 |
| Offset |
0 |
| Orientation |
Reverse Complement |
|
|
| Create custom LOGO ↧ |
[Next Match] [Query Top] |
Summary
|
Alignment
|
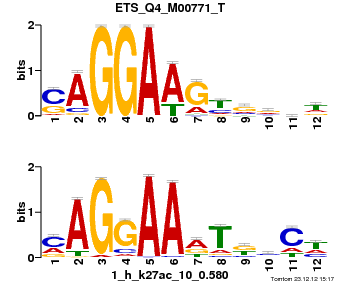
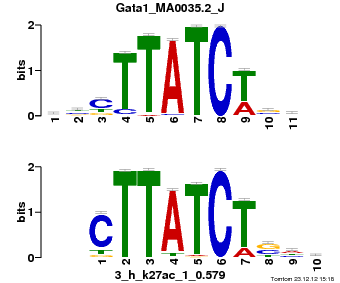
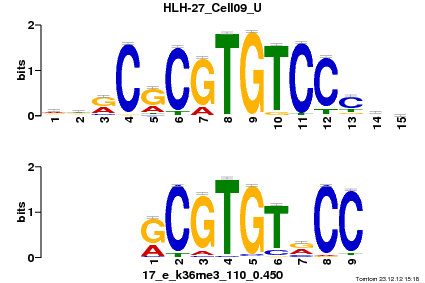
| Name |
Gata1_MA0035.2_J |
| Database |
trnsfcJsprUnprobHpdiCmbned3.meme |
|
p-value |
3.31838e-07 |
|
E-value |
0.000969298 |
|
q-value |
0.000966999 |
| Overlap |
9 |
| Offset |
2 |
| Orientation |
Reverse Complement |
|
|
| Create custom LOGO ↧ |
[Previous Match] [Next Match] [Query Top] |
Summary
|
Alignment
|
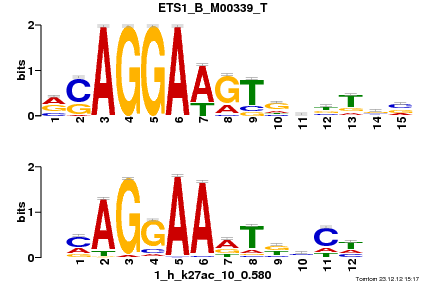
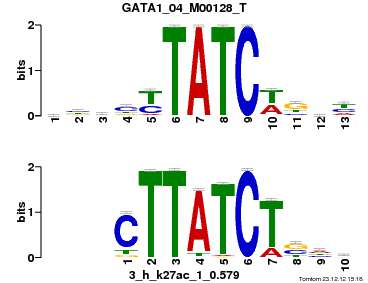
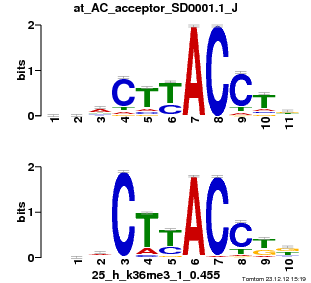
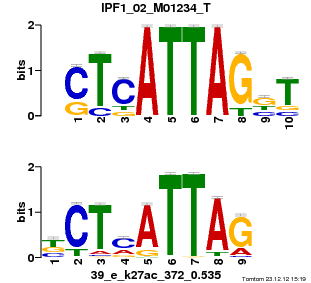
| Name |
GATA1_04_M00128_T |
| Database |
trnsfcJsprUnprobHpdiCmbned3.meme |
|
p-value |
1.63829e-06 |
|
E-value |
0.00478545 |
|
q-value |
0.00318273 |
| Overlap |
10 |
| Offset |
3 |
| Orientation |
Reverse Complement |
|
|
| Create custom LOGO ↧ |
[Previous Match] [Next Match] [Query Top] |
Summary
|
Alignment
|
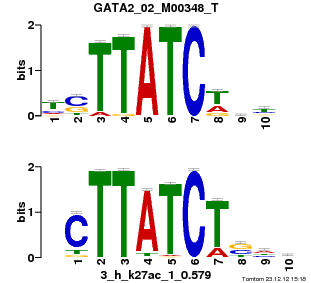
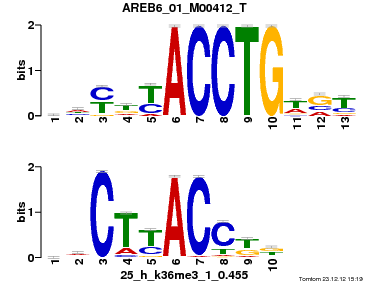
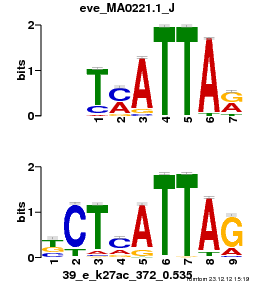
| Name |
GATA2_02_M00348_T |
| Database |
trnsfcJsprUnprobHpdiCmbned3.meme |
|
p-value |
3.14131e-06 |
|
E-value |
0.00917577 |
|
q-value |
0.004577 |
| Overlap |
9 |
| Offset |
1 |
| Orientation |
Reverse Complement |
|
|
| Create custom LOGO ↧ |
[Previous Match] [Next Match] [Query Top] |
Summary
|
Alignment
|
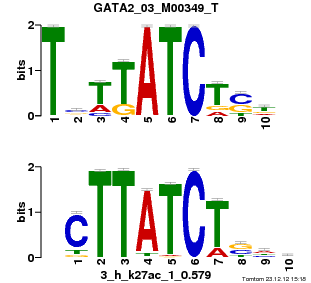
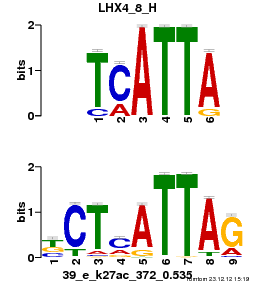
| Name |
GATA2_03_M00349_T |
| Database |
trnsfcJsprUnprobHpdiCmbned3.meme |
|
p-value |
5.76754e-06 |
|
E-value |
0.016847 |
|
q-value |
0.00611247 |
| Overlap |
9 |
| Offset |
1 |
| Orientation |
Reverse Complement |
|
|
| Create custom LOGO ↧ |
[Previous Match] [Next Match] [Query Top] |
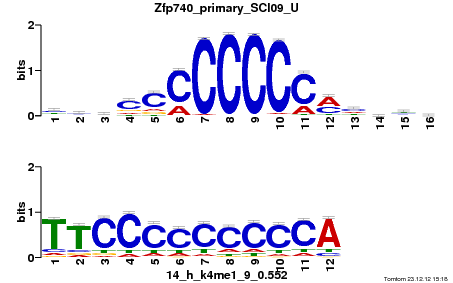
Summary
|
Alignment
|
| Name |
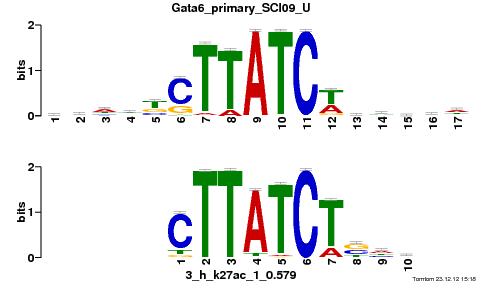
Gata6_primary_SCI09_U |
| Database |
trnsfcJsprUnprobHpdiCmbned3.meme |
|
p-value |
6.29271e-06 |
|
E-value |
0.018381 |
|
q-value |
0.00611247 |
| Overlap |
10 |
| Offset |
5 |
| Orientation |
Reverse Complement |
|
|
| Create custom LOGO ↧ |
[Previous Match] [Next Match] [Query Top] |
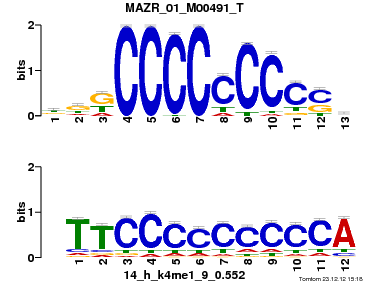
Summary
|
Alignment
|
| Name |
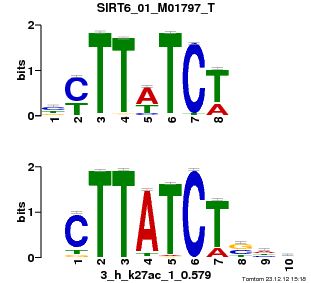
SIRT6_01_M01797_T |
| Database |
trnsfcJsprUnprobHpdiCmbned3.meme |
|
p-value |
1.22632e-05 |
|
E-value |
0.0358208 |
|
q-value |
0.0102102 |
| Overlap |
7 |
| Offset |
1 |
| Orientation |
Reverse Complement |
|
|
| Create custom LOGO ↧ |
[Previous Match] [Next Match] [Query Top] |
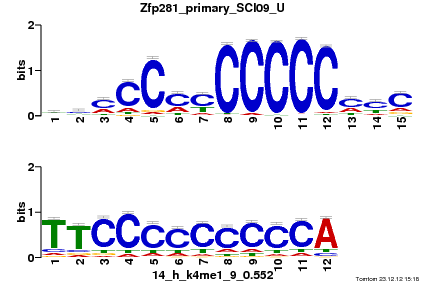
Summary
|
Alignment
|
| Name |
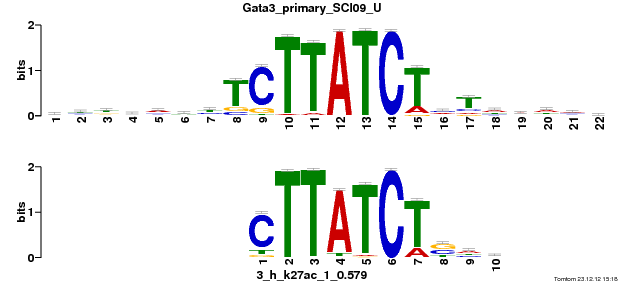
Gata3_primary_SCI09_U |
| Database |
trnsfcJsprUnprobHpdiCmbned3.meme |
|
p-value |
1.46516e-05 |
|
E-value |
0.0427973 |
|
q-value |
0.010674 |
| Overlap |
10 |
| Offset |
8 |
| Orientation |
Reverse Complement |
|
|
| Create custom LOGO ↧ |
[Previous Match] [Query Top] |