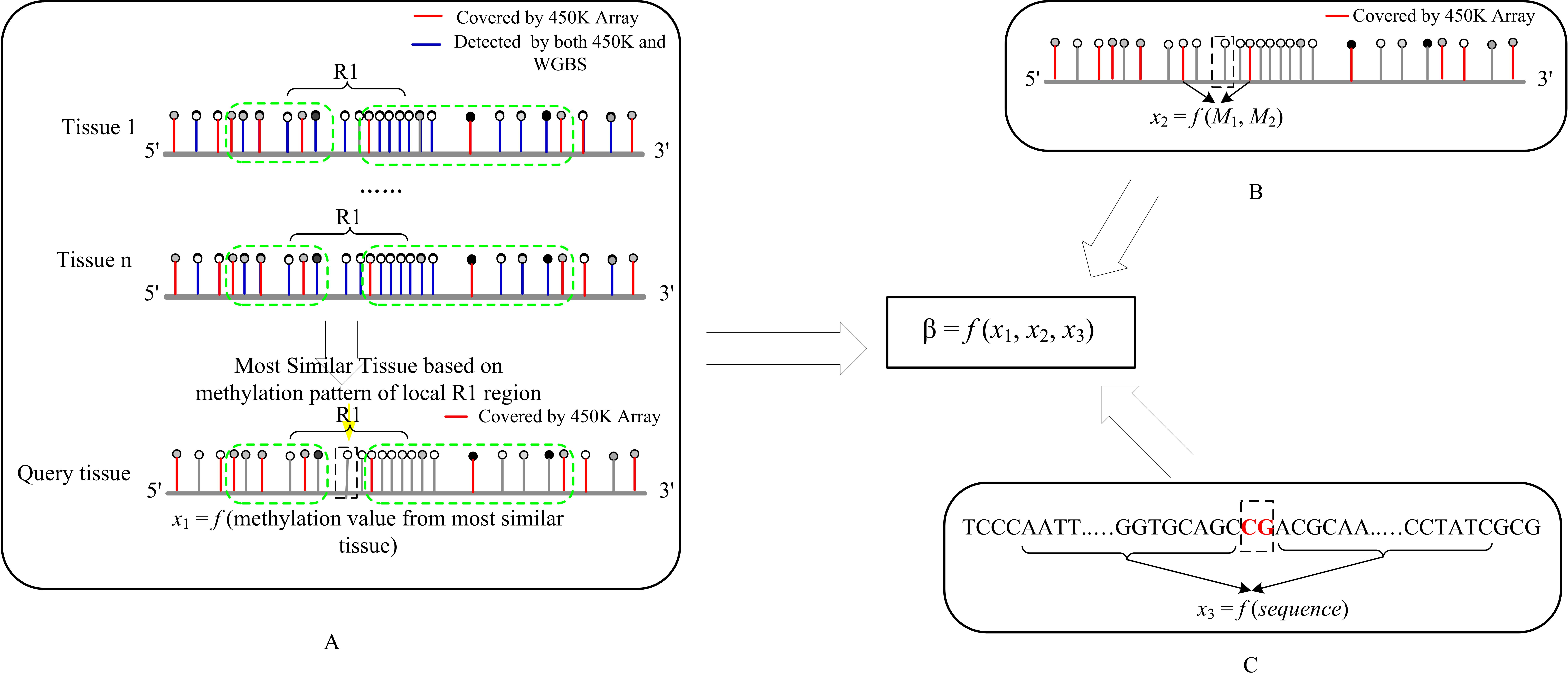
We developed a computational model that could expand the current 450K array data to a much broader scale. It provides a meaningful way to investigate the DNA methylome of various tissues, which are with 450K array data, but without histone modification data or DNA methylation data detected with other experimental techniques. This model integrates information derived from the similarity of local methylation pattern between tissues, the methylation information of flanking CpG sites and the methylation tendency of flanking DNA sequences. The main flowchart is as below.
The trained models for each chromosome can be downloaded here: ModelwithSeq or ModelwithoutSeq
The source code for prediction is available here: Source code in R
The data required for the code running is available here: Required Data
A test dataset is available here: Demo data for test
For instructions on how to use the code please go to the : Documentation
Please contact Wei Wang (wei-wang at ucsd.edu) for any question on our model.