EpiTensor: constructing 3-D interactions from 1-D epigenomes
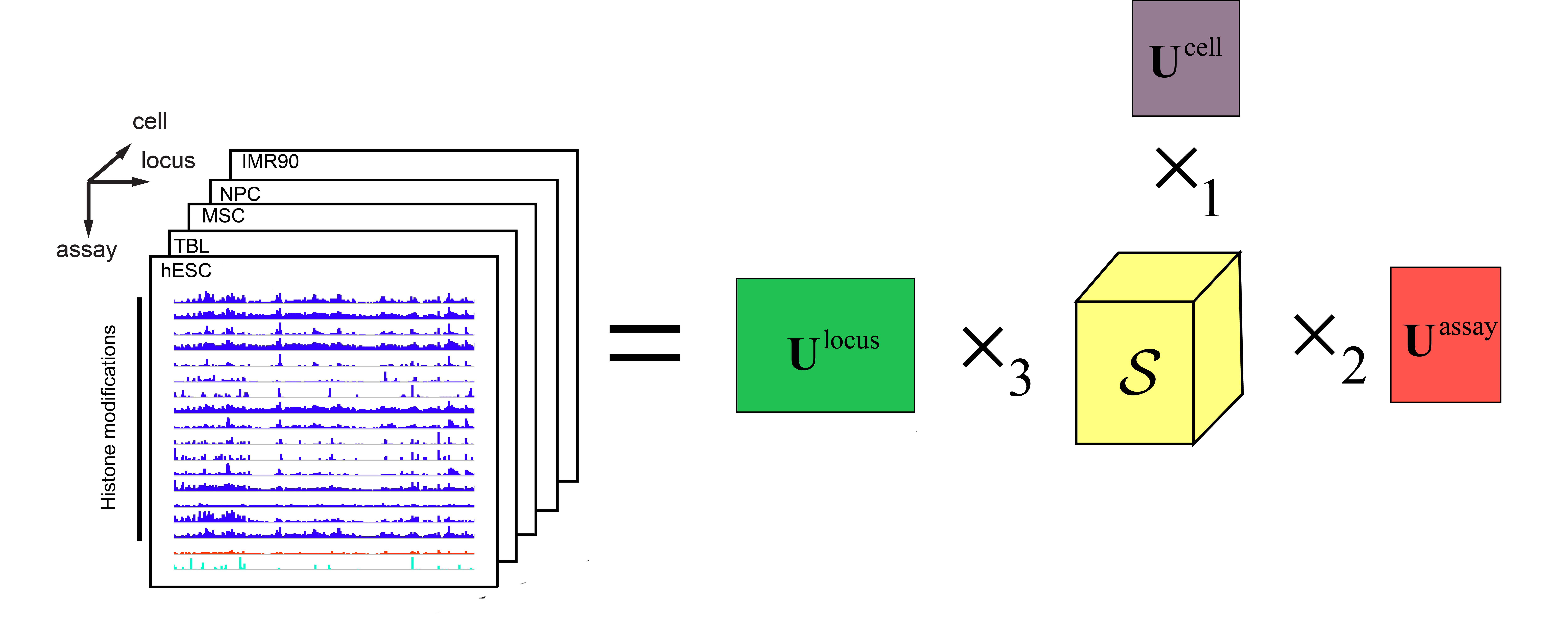 Epitensor
is a software package that can construct 3-D interactions from 1-D
epigenomes. It
uses tensor
to represent high-dimensional epigenomic data of various assays in
multiple cell types, and the utilizes tensor decomposition to break the
tensor into cell-type, assay, and genomic locus subspaces. The genomic
locus subspace captures epigenomic patterns across distal loci and
is associated with 3-D interactions among regulatory elements
Epitensor
is a software package that can construct 3-D interactions from 1-D
epigenomes. It
uses tensor
to represent high-dimensional epigenomic data of various assays in
multiple cell types, and the utilizes tensor decomposition to break the
tensor into cell-type, assay, and genomic locus subspaces. The genomic
locus subspace captures epigenomic patterns across distal loci and
is associated with 3-D interactions among regulatory elements
Download
Epitensor v0.9Documentation
Epitensor v0.9 manual
Quick Start
1. Installation (64-bit LINUX distribution):1) Unzip the downloaded Epitensor package using the following command
tar –zxvf epitensor.tar.gz to unzip the package.
The folder "release" should now be available.
2) Download MATLAB Compiler Runtime (MCR, http://www.mathworks.com/supportfiles/MCR_Runtime/R2013a/MCR_R2013a_glnxa64_installer.zip) use the following commands to install MCR:
./install
Alternatively, you can install MCR non-interactively using the following commands:
./install -mode silent -agreeToLicense yes -destinationFolder <MCRROOT>,where <MCRROOT> is the root directory of MCR (e.g. /home/yun/MATLAB/MATLAB_Compiler_Runtime/v81).
When the installation is done, add the following line to your .bashrc file:
export MCRROOT=<MCRROOT>,
3) Install R enviroment (http://www.r-project.org/)
The LINUX command "R" should now start the R environment
4) Once R is installed, type "R" in LINUX console to enter the R environment. Use "install.packages("R.matlab")" to install "R.matlab" package.
Try "library("R.matlab")", and it should work in R environment
5) Download spp package (http://compbio.med.harvard.edu/Supplements/ChIP-seq/).
Install Boost C++ if not already installed.
use "R CMD INSTALL spp_1.10.tar.gz" in linux console to install spp package
Try "library("spp")" and it should work in R environment
6) Download wigToBigWig (http://hgdownload.cse.ucsc.edu/admin/exe/linux.x86_64/). Only wigToBigWig is needed.
Edit your ~/.bashrc file to include:
PATH=$PATH:<KENT>, where <KENT> is the installation directory (e.g. PATH=$PATH:/home/yun/software/kent/)
The command wigToBigWig should now work from the command line.
7) Download MACS2 software (https://github.com/taoliu/MACS/wiki/Install-macs2) and follow the instructions to install it.
Try the "macs2" command in LINUX and macs2 usage instruction should pop out.
8) Download BEDTools (v2.17) (https://github.com/taoliu/MACS/wiki/Install-macs2).
EDIT your ~/.bashrc file to include:
PATH=$PATH; <BEDTOOLS>, where <BEDTOOLS> is the installation directory (e.g. PATH=$PATH:/home/yun/bedtools-2.17.0/bin/)
9) Use "sh recompile.sh" command to compile est_bkgd.lambda.cpp file.
Try the command "mergeBed -?" in LINUX and mergeBed usage instruction should pop out. The "-nms" should be listed as one of the parameters. Please note that BEDTools of version later than 2.20 will not work because the "-nms" parameter is not available.
2. Run Epitensor:
Try out "demo2.bash" to predict tss-tss, tss-enhancer, enhancer-enhancer interactions in H1, TBL, MSC, NPC, and IMR90 cells.
./demo2.bash
After temination in ~90 minutes, the "tss_tss_interaction.bed", "tss_enh_interaction.bed", and "enh_enh_interaction" will be available in the "out" directory.
The software package was tested on a workstation with Intel Xeon CPU and 12 GB memory.
Interaction Data
hESCIMR90
MSC
NPC
TRO